The genomic signature of sexual selection
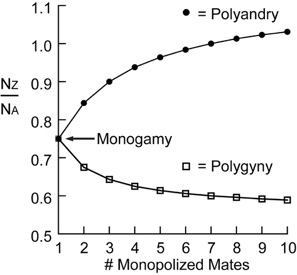
I have investigated how differences in the levels of sexual selection experienced by a species may affect the patterns of genetic diversity observed across the genome. Population genetics theory proposes that variance in male reproductive success will lower the effective population size of DNA predominantly transmitted by males. This will cause the sex chromosomes to exhibit different levels of genetic diversity relative to the autosomes (the Nz/Na ratio in the graph) under monogamous, polygynous, and polyandrous mating systems. To test this idea, I surveyed levels of genetic diversity in phylogenetically independent pairs of birds with different mating systems. I worked on shorebirds, which have exceptionally diverse mating systems that have long been of interest to biologists (the above illustration of a ruff is from Darwin’s book: The Descent of Man and Selection in Relation to Sex). I found that most polygynous species had relatively reduced genetic variation on their sex chromosomes as compared to monogamous species. These results suggest that measuring the differences in genetic diversity between the sex chromosomes and autosomes can contribute to understanding the long-term history of sexual selection experienced by a species. I did this work as a postdoc in the lab of Hans Ellegren at Uppsala University.
Publications
Corl, A., H. Ellegren. 2012. The genomic signature of sexual selection in the genetic diversity of the sex chromosomes and autosomes. Evolution. 66: 2138–2149. [link]