Sampling strategies for species trees
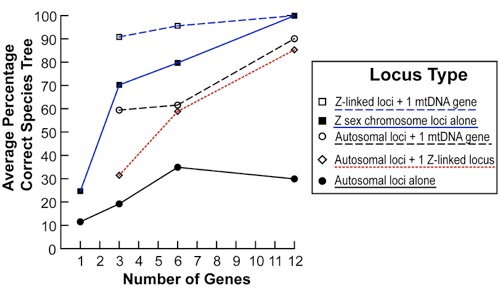
Systematists can now use multi-locus data to construct species trees that take into account the stochastic nature of gene tree divergence among populations. I evaluated how best to reconstruct a species tree in terms of numbers of genes, numbers of individuals per species, and types of loci. I studied sampling strategies using an empirical data set for six shorebird species in which I sequenced one mitochondrial, twelve autosomal, and twelve Z-linked loci for >8 individuals/species. I found that sampling greater numbers of genes resulted in substantial improvements to the resolution of the species tree, but sampling greater numbers of individuals had minor effects. I found that Z-linked loci significantly outperformed autosomal loci at all levels of sampling, which likely resulted from the lower effective population size of the Z-linked loci. Therefore, sex-linked loci are likely to be a powerful tool for multi-locus phylogenetic studies. I also found that adding a mitochondrial gene to a set of Z-linked or autosomal loci substantially improved the resolution of the tree. Overall, my results helped evaluate how best to maximize phylogenetic resolution while minimizing the costs of sequencing and computation when performing species tree analyses. I conducted this work as a postdoc in the lab of Hans Ellegren at Uppsala University.
Publications
Corl, A., and H. Ellegren. 2013. Sampling strategies for species trees: the effects on phylogenetic inference of the number of genes, number of individuals, and whether loci are mitochondrial, sex-linked, or autosomal. Molecular Phylogenetics and Evolution. 67: 358–366. [link]